Research Projects
This project, in collaboration with Jacqueline Cherfils,CNRS, France, aims to map, using high pressure coupled with fluorescence, NMR, SAXS and computational approaches, the nucleotide switch transitions for small Arf GTPases and characterize the molecular basis for their GEF/GAP specificity
Learn MoreThe precise coupling of cell growth and division is vital for the survival and fitness of all organisms. Division typically depends on cell growth, manifested as a homeostatic cell size that is characteristic for a given species, ploidy or tissue type. Disruption of the networks that control cell growth, division or size is linked to many disease phenotypes, including cancer, metabolic syndrome, and cardiomyopathy, amongst others. In budding yeast, the cell size threshold is tuned by nutrient conditions: in rich medium cells grow quickly and are large, whereas in poor medium they are small. In collaboration with the Tyers group at the IRIC at the University of Montreal we aim to decipher the molecular mechanisms controlling commitment to division (and hence size) in budding yeast. We use a combination of fluctuation microscopy, FLIM/Phasor and super-resolution single molecule localization SMLM) microscopy to quantitatively characterize the main transcription factors of the G1/S regulon.
Learn MoreThe global objective of the project, in collaboration with Doug Bartlett at UCSD and the Scripps Institute for Oceanography, is to reveal molecular mechanisms of transcriptional response and adaptation to high pressure using advanced microscopy techniques (scanning Number and Brightness, FLIM/Phasor) coupled with a high pressure, variable tempearture two-photon microscope
Learn MoreThis project, in collaboration with Doug Barrick and Bertrand Garcia-Moreno at Johns Hopkins University and Dan Raleigh at SUNY Stony Brook, aims to map, using high pressure coupled with fluorescence, NMR, SAXS and computational approaches, the folding free energy landscapes for selected model proteins. We demonstrated that pressure effects are based on the elimination of local packing defects in protein folded structures upon unfolding. Our current interests involve defining the sequence determinants of protein conformational landscapes.
Learn MoreThe global objective of the project, in collaboration with Roland Winter at TU-Dortmund, Jody Puglisi at Stanford and Eric Holden at Boise State, is to structurally and energetically characterize tertiary conformational transitions of structured RNA molecules using high hydrostatic pressure coupled to fluorescence, FTIR, SAXS and NMR. Pressure effects on biopolymer conformational equilibria are due to differences in molar volume between states, which for RNA, arise from differences in voids and hydration effects. Pressure perturbation of RNA allows quantitative assessment of the changes in hydration and ion condensation implicated in RNA structural dynamics...
Learn MoreIn collaboration with Nathalie Declerk and Abram Aertsen, we have investigated the pressure-induced response of the bacterial restriction endonuclease Mrr to high pressure using advanced microscopy techniques (scanning Number and Brightness, FLIM/Phasor)
Learn MoreMapping the nucleotide switch transitions in small G proteins (Arfs)
 Our overall goal is to reveal the molecular determinants underlying the GDP/GTP switches and functional specificities of Arf1 and Arf6 GTPases. Our approach combines pressure perturbation to populated excited states, with biophysical tools (multi-dimensional NMR, SAXS and fluorescence) and coarse-grained molecular dynamics simulations constrained by our data, to provide structural ensembles and pseudo-free energy landscapes. We are applying this approach to decipher the pathways of the GDP/GTP switch in the Arf GTPases and the physical mechanisms underlying their distinct specificity.
Our overall goal is to reveal the molecular determinants underlying the GDP/GTP switches and functional specificities of Arf1 and Arf6 GTPases. Our approach combines pressure perturbation to populated excited states, with biophysical tools (multi-dimensional NMR, SAXS and fluorescence) and coarse-grained molecular dynamics simulations constrained by our data, to provide structural ensembles and pseudo-free energy landscapes. We are applying this approach to decipher the pathways of the GDP/GTP switch in the Arf GTPases and the physical mechanisms underlying their distinct specificity.
Aim 1. Structurally and energetically map the GDP/GTP switch of Arf1. We hypothesize that the Arf GDP/GTP switch involves significant partial unfolding. We will map the Arf1 GDP- and GTP-bound conformational landscapes using our pressure-based mapping approach. The intermediate states thus characterized will be used to construct a structural and energetic model for the Arf1 GDP/GTP switch transition.
Aim 2. Structurally and energetically map the GDP/GTP switch of Arf6. We hypothesize that the Arf6 GDP/GTP switch transition involves even more highly disordered intermediates than Arf1. As in Aim 1, we will use pressure-based configurational mapping to characterize Arf6 excited states and based on these, propose a structural and energetic model for the Arf6 GDP/GTP switch transition.
Aim 3. Establish the underlying molecular mechanisms for Arf1/Arf6 functional specificity. To refine the models for Arf1/Arf6 GDP/GTP switches we will probe the effects of swapping Ser38 in Arf6 and the equivalent Ile42 in Arf1. This swap increases the nucleotide exchange rate in Arf1 and decreases the rate in Arf6. We hypothesize that these modifications will lead to corollary swaps in the Arf1/Arf6 conformational ensembles and switch pathways.
Molecular Mechanisms of Cell Size Homeostasis in Budding Yeast
The precise coupling of cell growth and division is vital for the survival and fitness of all organisms.  Division typically depends on cell growth, manifested as a homeostatic cell size that is characteristic for a given species, ploidy or tissue type. Disruption of the networks that control cell growth, division or size is linked to many disease phenotypes, including cancer, metabolic syndrome, and cardiomyopathy, amongst others. In budding yeast, the cell size threshold is tuned by nutrient conditions: in rich medium cells grow quickly and are large, whereas in poor medium, cells grow slowly and are small. Cells must somehow sense and set their size in order to maximize fitness in the face of ever-changing environmental conditions.
Division typically depends on cell growth, manifested as a homeostatic cell size that is characteristic for a given species, ploidy or tissue type. Disruption of the networks that control cell growth, division or size is linked to many disease phenotypes, including cancer, metabolic syndrome, and cardiomyopathy, amongst others. In budding yeast, the cell size threshold is tuned by nutrient conditions: in rich medium cells grow quickly and are large, whereas in poor medium, cells grow slowly and are small. Cells must somehow sense and set their size in order to maximize fitness in the face of ever-changing environmental conditions.
Despite the identification of literally hundreds of genes that affect size, how the size threshold is set and how size is sensed have defied a mechanistic understanding. Our project, in collaboration with the Tyers group at the IRIC in Montreal, involves quantitative mapping of the central molecular interactions involved in the commitment to division using advanced microscopy (2p sN&B, FLIM Phasor and PALM/STORM) in living yeast cells.
Publications:
Dorsey, S., Tollis, S., Cheng, J., Black, L., Notley, S., Tyers, M. and Royer, C. A. G1/S Transcription Factor Copy Number is a Growth-Dependent Determinant of Cell Cycle Commitment in Yeast, Cell Systems, 6, 539+, (2018).
Papini, C. & Royer, C.A., Scanning Number and Brightness Yields Absolute Protein Concentrations in Live Cells: A Crucial Parameter Controlling Functional Bio-molecular Interaction Networks, Biophysical Reviews 10, 87-96 (2018).
Black, L., Tollis, S., Fu, G., Fiche, J.-B., Dorsey, S., Cheng, J., Notley, S., Crevier, B., Bigness, J., Nollmann, M., Tyers, M. and Royer, C. A. G1/S transcription factors assemble in discrete clusters that increase in number as cells grow, J. Cell. Biol. 219, e202003041 (2020). 10.1083/jcb.202003041
Litsios, A., Goswami, P., Terpstra, H.M., Coffin, C., Vuillemenot, L.-A., Rovetta, M., Ghazal, G., Guerra, P., Buczac, K., Schmidt, A., Tollis, S. Tyers, M.T., Royer, C.A., Milas-Argeitis, A. & Heinemann, M., The timing of Start is determined primarily by increased synthesis of the Cln3 activator rather than dilution of the Whi5 inhibitor, Mol. Biol. Cell. (in press).
Tollis, S., Singh, J., Palou, R., Thattikota, Y., Ghazal, G., Coulombe-Huntington, J., Tang, X., Moore, S., Blake, D., Bonneil, E., Royer, C.A., Thibault, P. & Tyers, M., The microprotein Nrs1 rewires the G1/S transcriptional machinery during nitrogen limitation in budding yeast, PLoS Biology 20, e3001548 (2022).
Transcriptional adaptation to pressure
Transcriptional response to pressure
Among the cellular processes most affected by pressure are those implicated in the transcription. The objective of the present proposal is to reveal molecular mechanisms of transcriptional response and adaptation to high pressure. To achieve this objective, we will use fluorescent protein promoter and protein fusions and quantitative high-pressure advanced live single-cell microscopy approaches (scanning number and brightness and raster scanning image correlation) to measure absolute concentrations, dynamics and interactions of proteins involved in these transcriptional responses to pressure.
Publications:
Bourges, A., A. Lazarev, N. Declerck, K.L. Rogers, and C. Royer. 2020. Quantitative high-resolution imaging of live microbial cells at high hydrostatic pressure. Biophys. J. 118, 2670-2679 (2020).
Pressure Based Mapping of Protein Conformational Landscapes
This project, in collaboration with Doug Barrick at Johns Hopkins University and Dan Raleigh at SUNY Stony Brook, aims to map, using high pressure coupled with fluorescence, NMR, SAXS and computational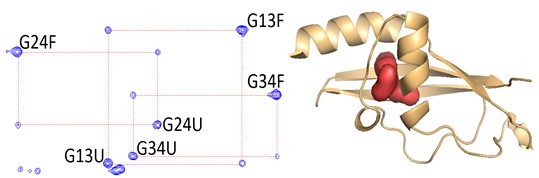 approaches, the folding free energy landscapes for selected model proteins.
approaches, the folding free energy landscapes for selected model proteins.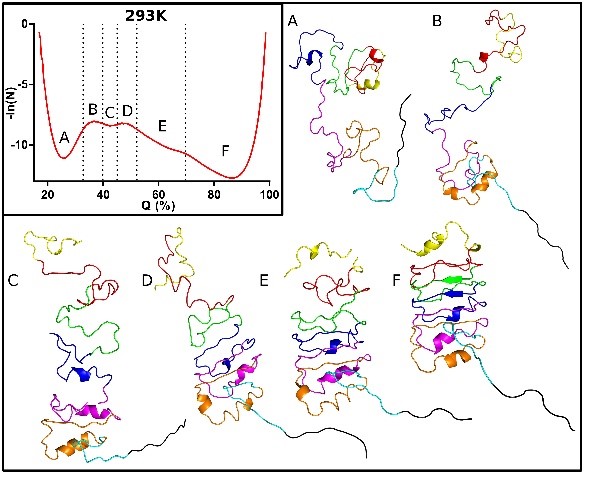 We seek to identify the sequence and structural determinants of their folding cooperativity, initiation and pathways. We recently demonstrated that pressure effects are based on the elimination of local packing defects in protein folded structures upon unfolding (Rouget et al 2012, Roche et al 2012). Thus, pressure perturbation is distinct from chemical denaturation, which is based on the amount of surface area exposed in unfolded states of proteins. The unique mechanism of pressure denaturation allows to more readily populate, and hence characterize, folding intermediates.
We seek to identify the sequence and structural determinants of their folding cooperativity, initiation and pathways. We recently demonstrated that pressure effects are based on the elimination of local packing defects in protein folded structures upon unfolding (Rouget et al 2012, Roche et al 2012). Thus, pressure perturbation is distinct from chemical denaturation, which is based on the amount of surface area exposed in unfolded states of proteins. The unique mechanism of pressure denaturation allows to more readily populate, and hence characterize, folding intermediates.
Publications:
Harish, B., Gillilan, R.E., Zou, J., Wang, J., Raleigh, D.P. & Royer, C. A. Protein unfolded states populated at high and ambient pressure are similarly compact, Biophys. J. 120, 2592-2598 (2021).
Zhang, S., Zhang, Y., Stensoski, N., Peran, I., McCallum, S.A., Raleigh D. & Royer, C.A. Pressure-Temperature Analysis of the Stability of the CTL9 Domain Reveals Hidden Intermediates Folding Intermediates of CTL9, Biophys. J. 116, 445-453 (2019).
Roche, J. & Royer, C. A. Lessons from Pressure Denaturation of Proteins, J. Royal Soc. Interface, (in press) 2018.
Jenkins, K.A., Fossat, M.J., Zhang, S., Rai, D.K., Klein, S. Gillilan, R., White, Z., Gerlich, G., McCallum, S.A., Winter, R., Gruner, S.M., Barrick, D.& Royer, C.A. Consequences of Cavities on the Folding Landscape of a Repeat Protein Depend Upon Context, Proc. Natl. Acad. Sci. USA 115(35):E8153-E8161 (2018).
Roche, J., Royer, C.A. & Roumestand, C., Exploring protein conformational landscapes using high pressure NMR, Methods Enz., (in press) (2018).
Zhang, Y., Berghaus, M., Klein, S., Jenkins, K., Zhang, S., McCallum, S. A., Morgan, J. E., Winter, R., Barrick, D. & Royer, C. A. High Pressure NMR and SAXS Reveals How Capping Modulates Folding Cooperativity of the pp32 Leucine Rich Repeat Protein, J. Mol. Biol. 430, 1336-1349 (2018).
Papini, C., Pandharapande, Royer, C. and Makhatadze, G. Putting the Piezolyte Hypothesis under Pressure, Biophys J. 113, 974-977 (2017).
Roche, J, Royer, C.A. & Roumestand, C. Monitoring Protein Folding Through High Pressure NMR Spectroscopy, Progress in Nuclear Magnetic Resonance Spectroscopy, (2017).
Kitazawa, S., Fossat, M.J., McCallum, S.A., Garcia, A.E and Royer, C.A. NMR and Computation Reveal a Pressure-Sensitive Folded Conformation of Trp-Cage, J. Phys. Chem B., 121, 1258-1267 (2017).
Zhang, Y., Kitazawa, S., Peran, I., Stenzoski, N., McCallum, S.A., Raleigh, D.P & Royer, C.A., High Pressure ZZ-Exchange NMR Reveals Key Features of Protein Folding Transition States, J. Am. Chem. Soc., 13, 15260–15266 (2016).
Fossat, M. J., Dao, T. P., Jenkins, K., Dellarole, M., Yang, Y., McCallum, S. A., Garcia, A. E., Barrick, D., Roumestand, C. and Royer, C. A. High-Resolution Mapping of a Repeat Protein Folding Free Energy Landscape, Biophys. J. 111, 2368-2376 (2016).
Dellarole, M., Caro, J.A., Roche, J., Fossat, M., Barthe, P., Garcia-Moreno E., B. Royer, C.A. and Roumestand, C. Evolutionarily conserved pattern of interactions in a protein revealed by local thermal expansion properties, J Am Chem Soc, 137, 9354-9362 (2015).
Sibille N, Dellarole M, Royer C, Roumestand C. Measuring residual dipolar couplings at high hydrostatic pressure: robustness of alignment media to high pressure, J Biomol NMR, 58, 9-16 (2014).
Roche J., Dellarole M., Caro J.A., Norberto D.R., Garcia A.E., Garcia-Moreno, E. B., Roumestand, C., Royer, C.A., Effect of Internal Cavities on Folding Rates and Routes Revealed by Real-time Pressure-Jump NMR Spectroscopy. J Am Chem Soc., 134, 14610-14618, (2013).
Dellarole, M., Kobayshi, K., Rouget, J.-B., Caro, J. A., Roche, J., Islam, M., Garcia-Moreno E., B., Kuroda, K. & Royer, C. A., Probing the Physical Determinants of Thermal Expansion of Folded Proteins, J. Phys. Chem. B. 117, 12742-9 (2013).
Dellarole, M., Roumestand, C., Royer, C. & Lecompte, J.T.J. Volumetric properties underlying ligand binding in a monomeric hemoglobin: A high pressure NMR study, Biochim. Biophys. Acta., 9, 1910-22 (2013).
Roche J, Caro JA, Dellarole M, Guca E, Royer CA, E BG, Garcia AE, Roumestand C. Structural, energetic and dynamic responses of the native state ensemble of staphylococcal nuclease to cavity-creating mutations. Proteins. 81, 1069-80. (2013).
Roche J, Dellarole M, Caro JA, Guca E, Norberto DR, Yang Y, Garcia AE, Roumestand C, García-Moreno B, Royer CA. Remodeling of the folding free energy landscape of staphylococcal nuclease by cavity-creating mutations. Biochemistry. 51, 9535-46 (2012).
Roche, J., Caro, JJ.A., Norberto D. R., Barthe, P., Roumestand, C., Schlessmann, J.L., Garcia, A.E., Garcia-Moreno E., B. & Royer, C.A. Cavities determine the pressure unfolding of proteins, Proc. Natl. Acad. Sci. USA 109, 6945-6950 (2012).
Rouget, J.-B., Aksel, T., Roche, J., Saldana, J.-L., Garcia, A. E., Barrick, D., and Royer, C. A. Size and sequence and the volume change of protein folding, J Am Chem Soc S, 133, 6020-7, (2011).
Kitahara, R., Hata, K., Maeno, A., Akasaka, K., Chimenti, M., Garcia-Moreno E., B., Schroer , M. A., Jeworrek, C., Tolan, M., Winter, R., Roche, J., Roumestand, C., Montet de Guillen, K. and Royer, C. A. Structural plasticity of staphylococcal nuclease probed by perturbation by pressure and pH, Proteins, 79, 1293-1305 (2011).
Schroer, M., Paulus, M., Jeworrek, C., Krywka, C., Schmake, S., Zhai, Y., Florian Weiland, D. C., Sahle, C., J., Chimenti, M., Royer, C. A., Garcia-Moreno, B., Tolan, M. and Winter, R. High pressure SAXS studies of folded and unfolded ensembles of proteins, Biophys. J., 99, 3430-3437 (2010).
Effects of Pressure on RNA Conformation
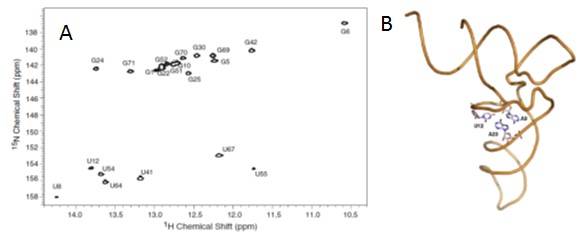 The global objective of the project, in collaboration with Jody Puglisi at Stanfrod and Eric Holden at Boise State, is to structurally and energetically characterize tertiary conformational transitions of structured RNA molecules using high hydrostatic pressure coupled to fluorescence, FTIR, SAXS and NMR.
The global objective of the project, in collaboration with Jody Puglisi at Stanfrod and Eric Holden at Boise State, is to structurally and energetically characterize tertiary conformational transitions of structured RNA molecules using high hydrostatic pressure coupled to fluorescence, FTIR, SAXS and NMR.
Pressure effects on biopolymer conformational equilibria are due to differences in molar volume between states, which for RNA, arise from differences in voids and hydration effects. Pressure perturbation of RNA allows quantitative assessment of the changes in hydration and ion condensation implicated in RNA structural dynamics.
Publications:
Gao, M., Harish, B., Berghaus, M., Seymen, R., Arns, L., McCallum, S., Royer, C. & Winter, R. Temperature and pressure limits of guanosine monophosphate self-assemblies, Scientific Rep. 7 (2017).
Knop, J.-M., Harish, B. Patra, S., Royer, C.A. & Winter, R., The Deep Sea Osmolyte TMAO and Macromolecular Crowders Rescue the Antiparallel Conformation of the Human Telomeric G-Quadruplex from Urea and Pressure Stress, Chemistry - A European Journal 24, 14346-14351 (2018).
Harish, B., Wang, J., Hayden, E.J., Grabe, B., Hiller, W, Winter, R. & Royer, C.A., Hidden intermediates in Mango III RNA aptamer folding revealed by pressure perturbation, Bophys. J., 121, 421-429 (2022).
Pressure-induced SOS response
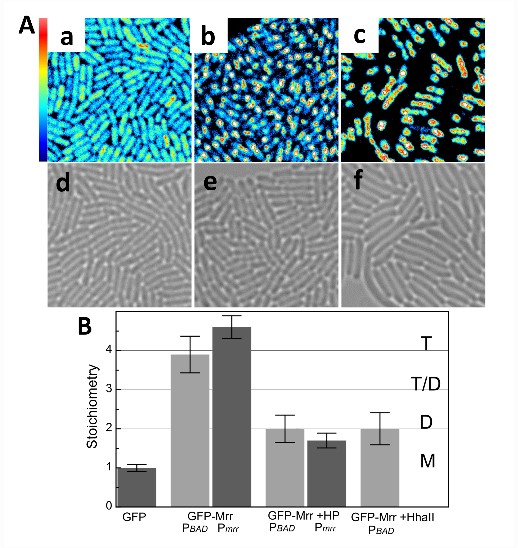 Prior screens for HP resistance identified Mrr, a Type IV restriction endonuclease (REase), as instigator for this enigmatic HP-induced SOS response. In a collaboration with Nathalie Declerck of the Center for Biochemistry in Montpellier, France and Abram Aertsen at the Katholieke University in Leuven our group uses high pressure coupled with 2-p sN&B to investigate the moelcular mechanisms underlying the the pressure-indced SOS response. Pressure and treatment by the HhaII methyltransferase induces irreversible formation of Mrr-GFP foci, indicative of the SOS response.
Prior screens for HP resistance identified Mrr, a Type IV restriction endonuclease (REase), as instigator for this enigmatic HP-induced SOS response. In a collaboration with Nathalie Declerck of the Center for Biochemistry in Montpellier, France and Abram Aertsen at the Katholieke University in Leuven our group uses high pressure coupled with 2-p sN&B to investigate the moelcular mechanisms underlying the the pressure-indced SOS response. Pressure and treatment by the HhaII methyltransferase induces irreversible formation of Mrr-GFP foci, indicative of the SOS response.
We are also investigating the effect of pressure on genome organization in bacteria. Below are images of the nucleoide protein Hu-mCherry and ParB-YFP,  which partitions plasmids into daughter cells. Images are taken at atmospheric pressure, 100 MPa (~ 1000 atmospheres) and after return t o atmospheric pressure. They show that in cells that survive pressurizaiton the LLPS foci made up of ParB-YFP are disrupted reversibly by pressure.
which partitions plasmids into daughter cells. Images are taken at atmospheric pressure, 100 MPa (~ 1000 atmospheres) and after return t o atmospheric pressure. They show that in cells that survive pressurizaiton the LLPS foci made up of ParB-YFP are disrupted reversibly by pressure.
Publications:
Bourges, A., Torres Montaguth, O.E., Ghosh, A., Tadesse, W.M., Declerck, N., Aertsen, A. and Royer, C.A. High pressure activation of the Mrr restriction endonuclease in Escherichia coli involves tetramer dissociation, Nuc. Acids. Res. 45, 5232-5332 (2017).
Declerck, N. & Royer, C.A. Interactions in gene expression networks studied by two-photon fluorescence fluctuation spectroscopy, in Fluctuation Spectroscopy, Methods in Enzymology, S. Tetin, ed., Springer, NY 519, 203-230 (2013).
Bourges A.C., Torres-Montaguth, O.E., Tadess, W., Labesse, G., Aertsen, A., Royer, C.A & Declerck, N. An oligomeric switch controls the Mrr-induced SOS Response in E. coli, DNA Repair, (in press) 2020.
Bourges, A., A. Lazarev, N. Declerck, K.L. Rogers, and C. Royer. 2020. Quantitative high-resolution imaging of live microbial cells at high hydrostatic pressure. Biophys. J. 118, 2670-2679 (2020).